Here's a quick overview of the different stages of the process I went through to make the 3D printed T-cell receptors detailed in
part 1.
Part 2: the process
Now there's a couple of nice tutorials kicking around on how to 3D print your favourite protein. One option which clearly has beautiful results is to use
UCSF's Chimera software, which was also the approach taken by
the first protein-printing I saw. However this seemed a little full-on for my first attempts, so I opted for relatively easy approach based on PyMol, modelling software I'm already familiar with.
The technique I used was covered very well in a
blog post by Jessica Polka, which was then covered again in a very well written
instructable. As these are both very nice resources I won't spend too long going over the same ground, but I thought it would be nice to fill in some of the gaps, maybe add a few pictures.
1: Find a protein
This should be the easy bit for most researchers (although if you work on a popular molecule finding the best crystal structure might take a little longer). Have a browse of the
Protein Data Bank, download the PDB and
open the molecule in PyMol.
All you need to do here is hide everything, show the surface, and export as a .wrl file (by saving as VRML 2). I mean that's all you
need to do. If you want to colour it in, that's totally fine too.
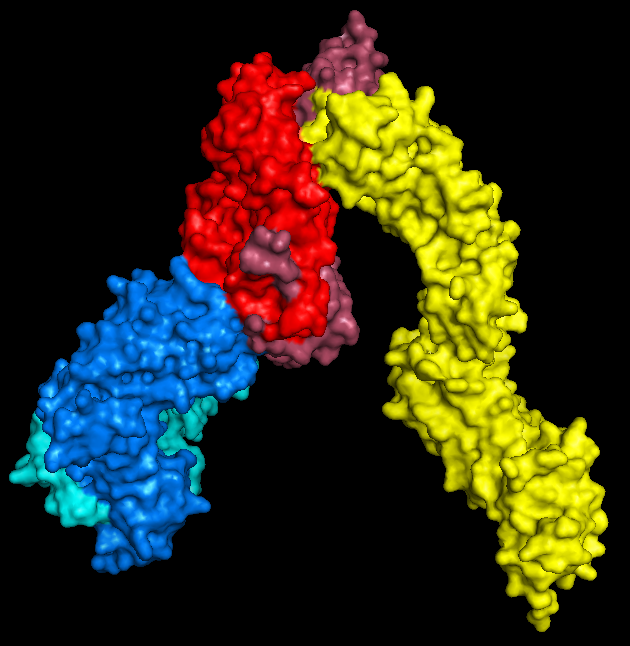 |
| PyMol keeps me entertained for hours. |
2: Convert to .stl
Nice and easy; open your .wrl file in
MeshLab ('Import Mesh'), and then export as a
.stl, which is a very common filetype for 3D structures.
 |
| Say goodbye to your lovely colours. |
3: Make it printable
Now we have a format that the proprietary 3D printing software can handle. As I primarily only have access to
MakerBots, I next open my stl files in
MakerWare.
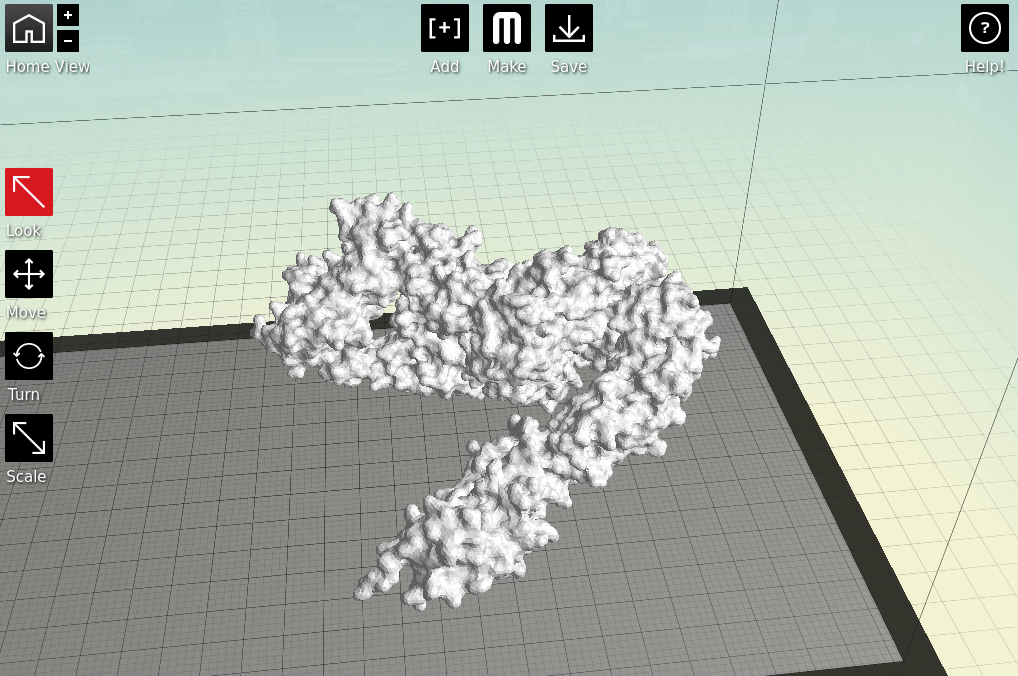 |
| Starting to feel real now, right? |
There's a couple of things to take into consideration here. First, is placement; you need to decide which side of your molecule will be the 'top'; remember, most protein structures are going to require scaffolds in order to be printed, which might cause some damage when removed.
Next is the quality of the print. One factor is the scale; the bigger you make your molecule the better it will look, and likely print, at the cost of speed and plastic. Additionally you can alter the thickness of each print layer, the percentage infill (how solid the inside of the print is, up to a completely solid print) and the number of 'shells' that the print has.
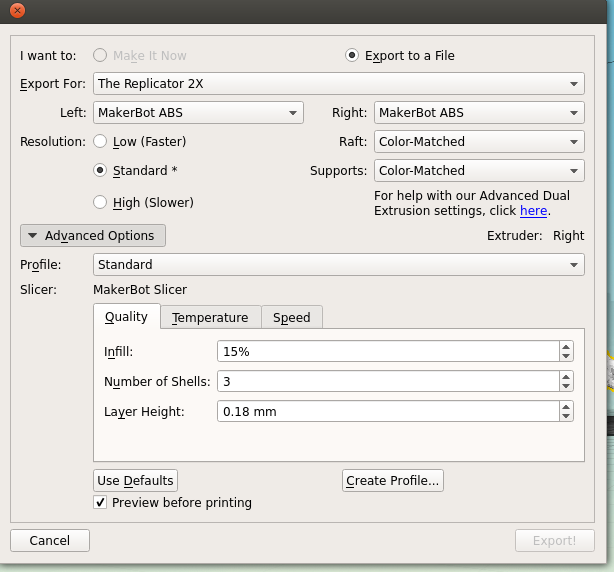 |
| Remember to tick 'Preview before printing' in order to get a time estimate. |
4: The print!
Both of my molecules so far have been printed on
MakerBot Replicator 2Xs, using
ABS plastic, taking between 10 and 14 hours per print due to the complexity and size of the models. This part is also nice and simple; just warm up your printer, load your file and click go.
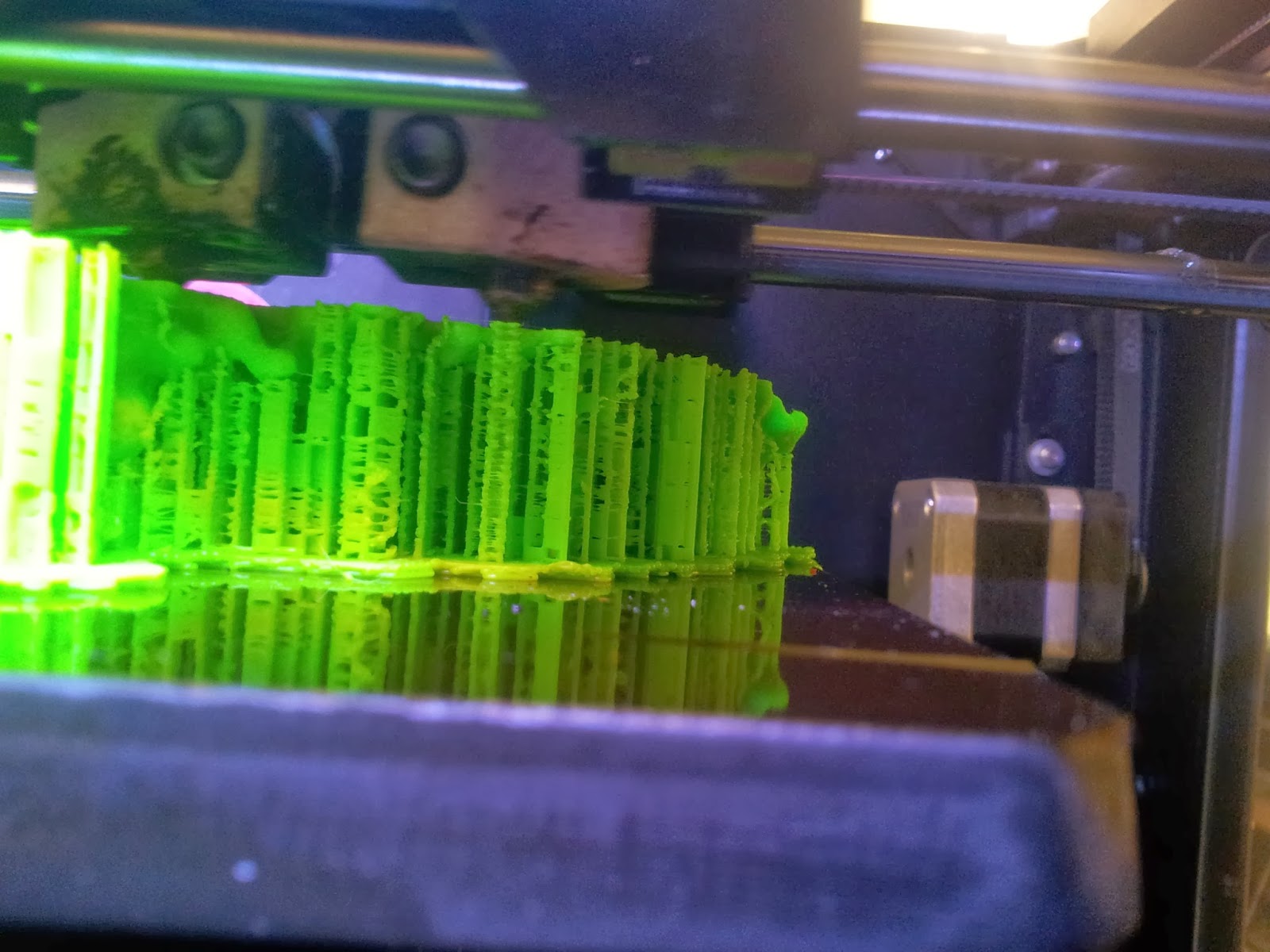 |
| A side view of the printer as it runs, detailing the raft and scaffolds that will support the various overhangs. |
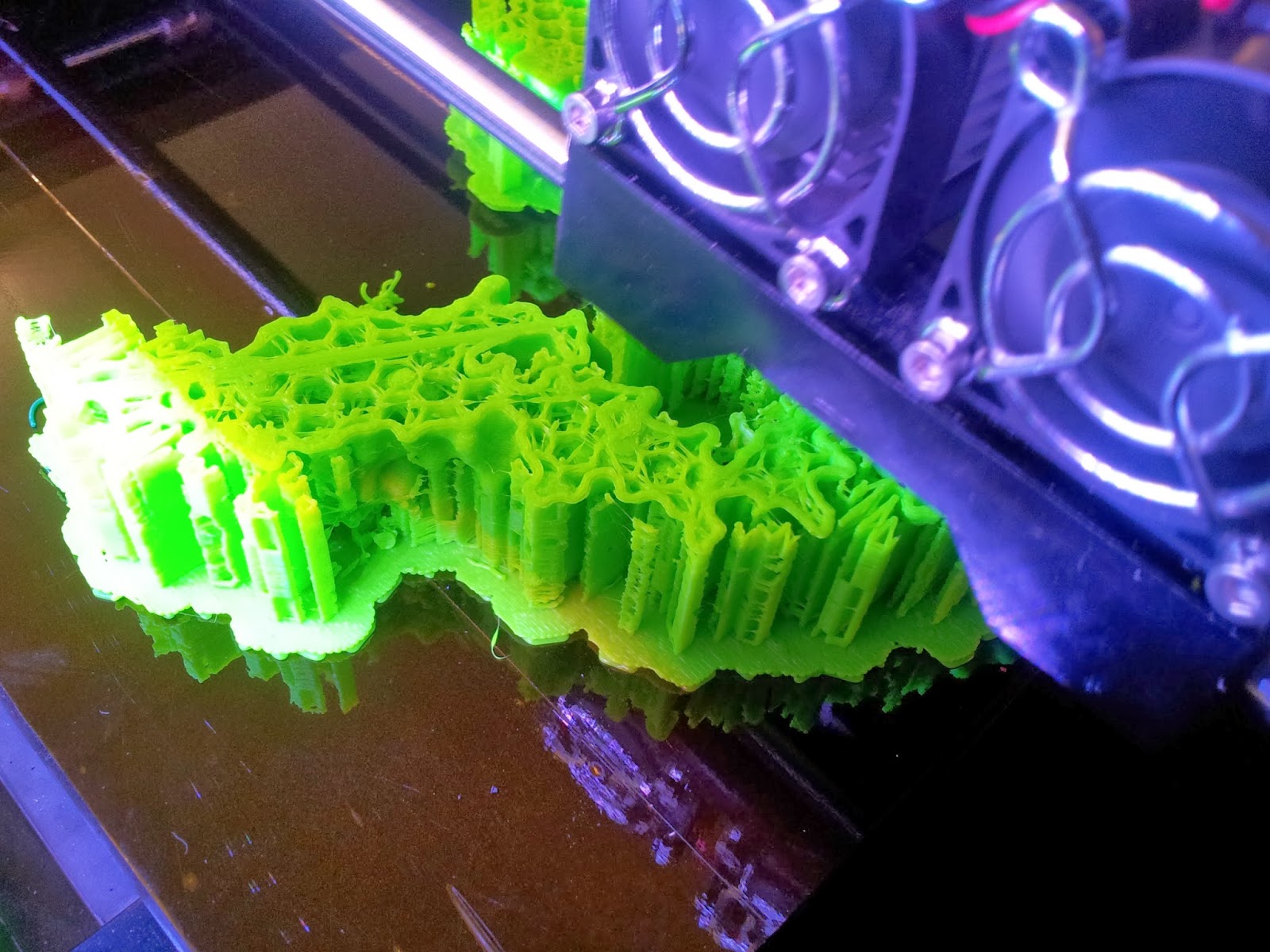 |
| The TCR begins to emerge, with hexagonal infill also visible. |
5: The tidy-up
The prints come out looking a little something like this:
 |
| Note the colour change where one roll of ABS ran out, and someone thoughtfully swapped it over, if sadly not for the same colour |
 |
| This green really does not photograph well. I like to pretend I was going for the nearest I could to the IMGT-approved shade of variable-region green, but really I was just picking the roll with the most ABS left to be sure it wouldn't run out again. |
Then comes the wonderfully satisfying job of ripping off the raft and the scaffolds. Words can't describe just how enjoyable (if incredibly fiddly and pokey) this is.
 |
| My thanks go to Katharine and Mattia for de-scaffolding services rendered. |
Seriously, this stuff is better than bubble wrap.
 |
| Prepare for mess and you will not be disappointed. Instead, you will be picking tiny bits out plastic out of your hair. |
I found a sliding scale of using needle-nose pliers, then tweezers, then very fine forceps seemed to work best. At this point make sure you keep some of your scrap ABS in reserve, as it can be useful later.
Once you've gotten all the scaffolding off, your protein should look the right shape, if a little scraggy around the edges. I've read that 3D printing people generally sometimes use fine sandpaper here to neaten up some of these edges, which I will consider in future, but generally the surface area to cover is fairly large and inaccessible, so it's not an option I've spent long dwelling on.
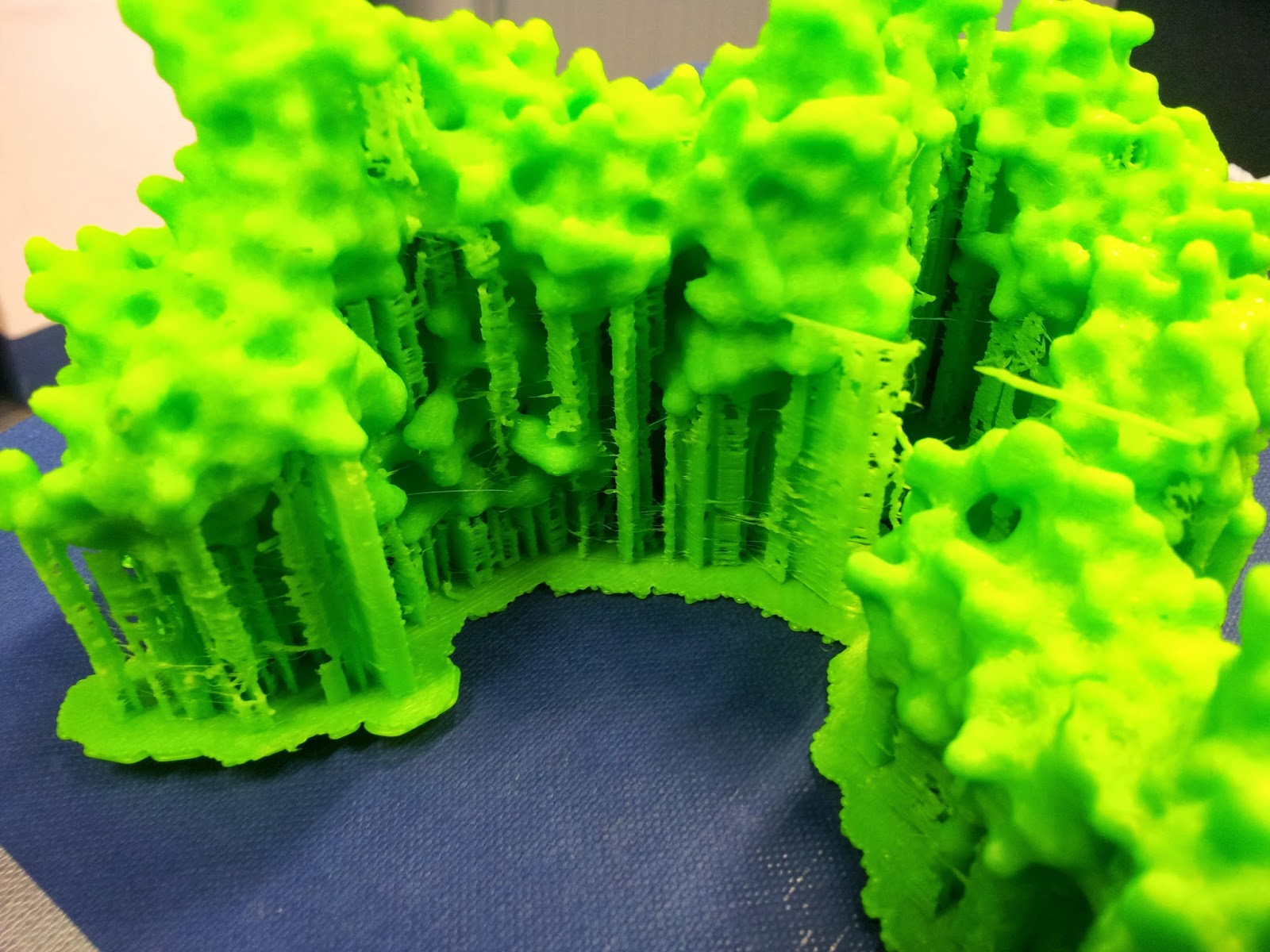
 |
| The nasty underbelly of the print, after scaffold removal |
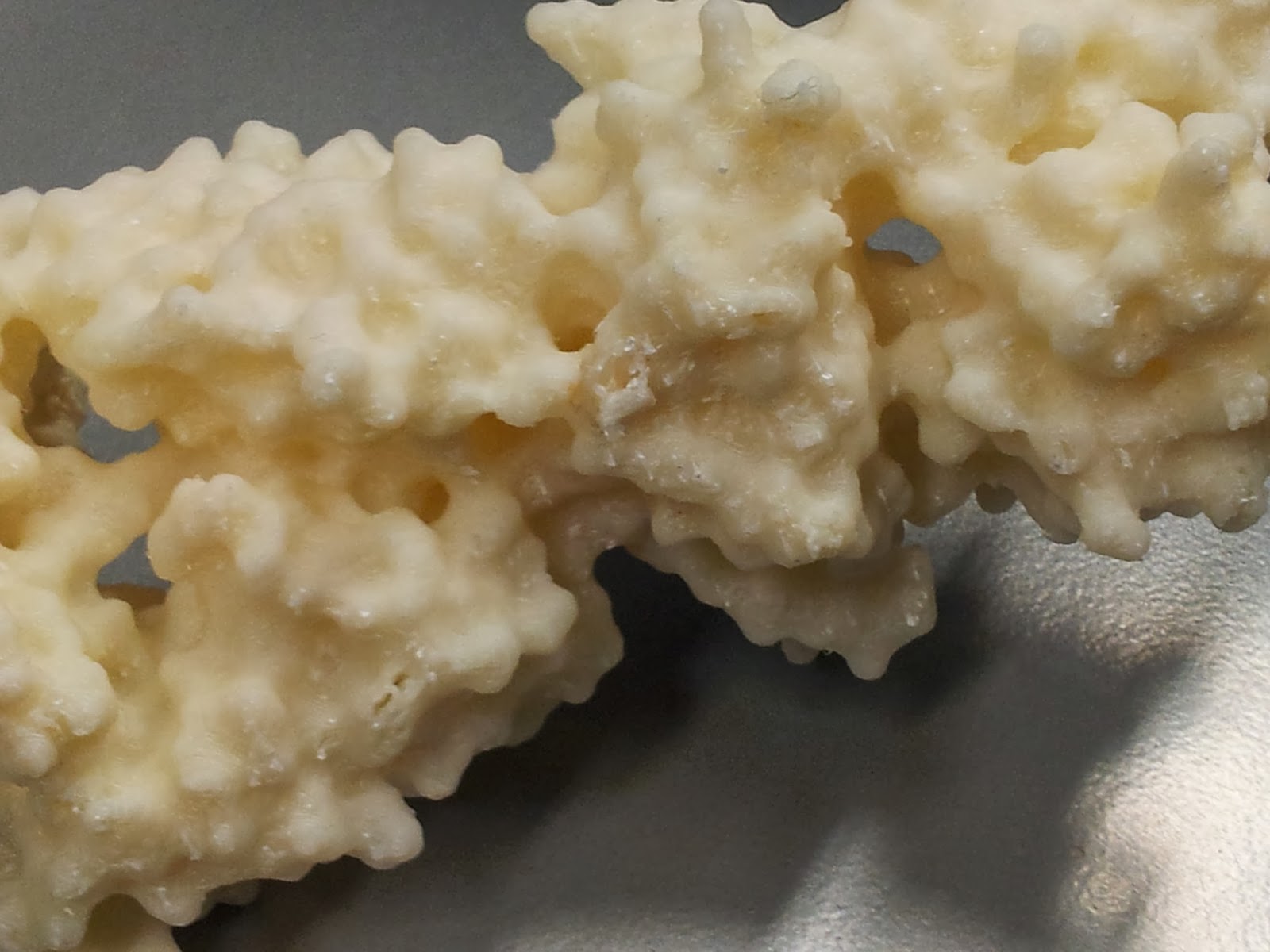
In an effort to minimise such unsightly bottoms in the second print, I went for a higher quality print than I had before (see above MakerWare dialog screenshot), however it still produced both scaffold bobbles and misprint holes - you can see two in the photo below, one just above and one just below slightly off centre to the right.
 |
| The other side is much nicer, I promise. |
 |
| Mmm, radioactive. |
However increasing the infill percentage and number of shells had one major noticeable difference; the side chains that stick out are much less fragile than they were on the first print*.
Note that this is also when the spare ABS can come in handy; dissolved in a little acetone, it readily becomes a sloppy paint, which can be slathered on to fill in any glaring flaws in the model.
I should point out that at this point the rest of the model tends to look pretty good (if I do say so myself).
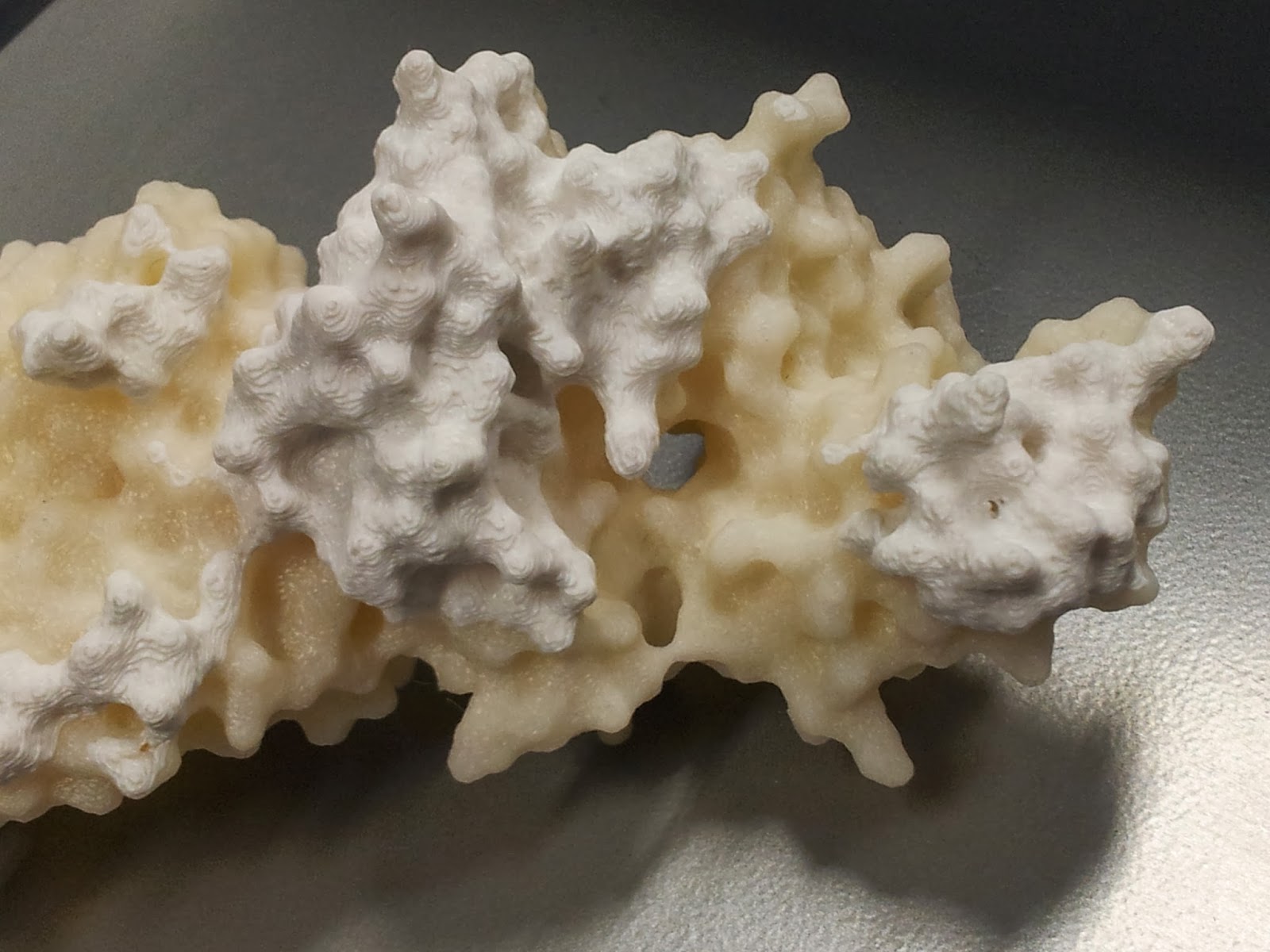 |
I got a lot of questions asking about the significance of the other colour.
|
6: Smoothing
In addition to physical removal of lumps, it's also possible to smooth out the printing layers themselves by exposing the print to acetone vapour for a time, as discussed in many nice
tutorials.
I personally like the contour effect of the printing somewhat, and due to the delicate nature of the protrusion-heavy proteins I didn't want to go for an extreme acetone shower, but I think a light application has smoothed off some of the rougher imperfections.
This is a particularly easy thing to achieve for the average wet-lab biologist, as we have ready access to heat blocks and (usually) large beakers. Unfortunately the 3T0E, CD4 containing print was too large for any of the beakers in the lab, so I had to make do.
 |
| Nothing pleases me more than a satisfactory bodge-job. |
I'm not sure if it was the larger volume or the denser or perhaps different plastic, but this set up took a lot longer to achieve lesser results than the previous model did. However it did still achieve a smoothing of the layers.
 |
| Still shiny - and tacky - from the acetone. |
It's worth doing this in a well ventilated area, as the combination of acetone vapour and melted-plastic smell aren't the nicest. Bear in mind the print itself will smell for a little while after smoothing, which will upset your office mates.
 |
| The finished
result; the model that made the immunologists of twitter all want rice
pudding. What a shame the nice side had the colour change. That the
acetone-smoothing appears to have affected the two colours differently
suggests that different rolls of ABS do indeed have different dissolving properties. |
There you have it, the simple method to easily 3D print the structure of proteins. Honestly, the hardest bit is finding a 3D printer to use.
Or in my case, finding the time to get over there to use it.
* I tell people that I was experimenting with tactile mutational analysis, when really I just dropped the print and a couple of aromatic side chains fell off. Note that they do readily stick back on with superglue.





















